Gene Mapping: Definition, Overview, Types, Techniques, Topics
Gene Mapping is a process of identifying the location of genes on a chromosome. It helps scientists to understand the exact position of specific genes. It provides a clear picture of how genes are arranged in DNA. It is an important tool for studying Human Genetic Disorders and inheritance patterns. It helps in understanding the Principle of Inheritance and Variations by locating genes responsible for traits.
This Story also Contains
- What Is Gene Mapping?
- Types Of Gene Mapping
- Linkage Mapping
- Physical Mapping
- Techniques Of Gene Mapping
- Applications Of Gene Mapping
- The Video Recommended On Gene Mapping:
- MCQs on Gene Mapping

By creating genetic maps, scientists can easily track the Inheritance pattern. It helps trace how genetic variations arose and spread within a population. Mapping genes is crucial to studying types of mutations and their role in evolution and diversity. Gene mapping helps identify gene interaction patterns that affect the inheritance and variation of complex traits. Genetically, these are specific genes or genetic markers that would help in navigation across the large landscape that a genome is.
What Is Gene Mapping?
Gene mapping is a central process in genetics that deals with the identification of specific locations of genes on a chromosome. This complex branch of study facilitates a detailed study of the arrangement and functions of genes within the DNA, also referred to as a blueprint of the genetic landscape. With the exact positions of genes known, one can further study their interaction with other genes and the environment to derive important discoveries in biology and medicine.
Gene mapping is of immense importance, as it forms the basis for many genetic studies and applications. Linkage and recombination help map gene positions on chromosomes. Gene mapping has changed everything about our perception relating to the human health system. Gene mapping in agriculture helps to improve plant and animal species by determining the richest sources of genes carrying desirable traits, such as resistance to diseases or higher yields.
Types Of Gene Mapping
In general, gene mapping could be broadly divided into a few types with their methodologies and applications. The main types of gene mapping include linkage mapping, physical mapping, and comparative mapping. Each type offers different levels of detail for studying the genome.
Linkage Mapping: It shows the position of genes by studying inheritance patterns. This type of mapping is based on the fact that genes already known to be physically close to one another will tend to be inherited together.
Physical Mapping: It finds the exact distance between genes on the DNA. It provides high-resolution maps of the genome.
Comparative Mapping: It compares genes of different species to find similarities. It may bring out evolutionary relationships and functional genomics.
Linkage Mapping
It is the mapping process that determines the relative positions of genes on a chromosome. It is based on the frequency of recombination between them.
It uses the process of recombination that goes forward during the stages of meiosis.
It is the kind of cell division by which gametes, sperm, and eggs are produced.
It is, therefore, less likely to have genes separated by recombination if they are physically close to each other on the same chromosome.
Genes farther apart have a higher probability of being recombined.
The major method applied in linkage mapping is constructing genetic linkage maps.
The construction of these maps is based on the crossing of individuals having different traits and the analysis of individual combinations of traits within the offspring.
The frequency of recombination between markers, either observable traits or DNA sequences themselves, is scored; data is then used in estimating gene distances.
Unit of Measurement: Centimorgan (cM)
The distance between genes is measured in centimorgans (cM).
1 centimorgan = 1% probability that recombination will separate two genes during crossing over.
Example: The map distance between genes A and B is 3 units, between B and C 10 units, and between C and A 7 units. The order of the genes in a linkage map is constructed:
Genes B and C would be at the extreme as they have a greater distance between them. Gene A would be in the middle position.
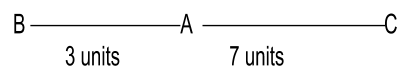
Example of linkage mapping
Linkage mapping has been used to find genes responsible for inherited diseases in humans. Researchers can trace a disease gene along with genetic markers in families having patients affected by a specific human genetic disorder.
Cystic fibrosis
Huntington's disease
Physical Mapping
It provides a more accurate and direct way of determining gene locations on a chromosome. In contrast to the linkage map, which is based on the frequencies of recombinations, physical mapping measures the actual distances between genes in base pairs. It involves breaking down the DNA into smaller fragments, sequencing them, and then assembling the sequences to create a complete map of the chromosome.
Different techniques of physical mapping
Restriction Mapping: DNA is cut with particular enzymes, and the lengths of the resulting fragments are used to estimate the distances between restriction sites.
FISH (Fluorescent In Situ Hybridization): It relies on the use of fluorescent probes that hybridise to target DNA sequences, allowing visual screening of the position of genes on the chromosomes via a microscope.
Sequencing-based approaches: These methods determine the entire DNA sequence of a genome. Example: Human Genome Project (HGP).
Techniques Of Gene Mapping
Gene mapping uses different techniques to identify the location of genes on chromosomes. These techniques help in both linkage mapping and physical mapping.
Restriction Fragment Length Polymorphism (RFLP): It is the technique that identifies changes in DNA sequence by monitoring the size of DNA fragments obtained due to restriction enzymes. This has been a powerful tool for linkage mapping and early genomic work, playing its role in mapping genetic markers related to diseases and traits.
Microsatellite Markers: Microsatellites are very small tandem repeats of DNA sequences, highly polymorphic, and distributed throughout genomes. They show a high degree of polymorphism, thus proving useful as genetic markers in linkage and physical mapping. It is the identification of genes and their location on chromosomes.
Single Nucleotide Polymorphisms (SNPs): Single base pair variation in DNA sequence is relatively common in populations. It is considered an important marker in gene mapping due to its frequency and association with diseases or traits. Identification and genomic mapping of SNPs are done with techniques like PCR-based assays and sequencing.
Fluorescence in Situ Hybridisation (FISH): It is a cytogenetic technique that involves the hybridisation of fluorescent probes to specific DNA sequences of chromosomes. It visualises directly, on chromosomes, the place where genes and genetic markers are being considered. Therefore, it gives spatial information about genome organisation in physical mapping.
Applications Of Gene Mapping
Gene mapping is the process of finding the location of genes on a chromosome. It helps scientists understand where specific genes are and how they are inherited. This knowledge is useful in medicine, agriculture, and studying evolution. It also helps in identifying genes linked to diseases or useful traits.
Field | Application of Gene Mapping |
Medical Genetics | Identifies disease-causing genes, helps in genetic testing and personalised medicine |
Agriculture | Improves crop yield, disease resistance, and livestock productivity |
Evolutionary Biology | Studies the evolution links, genetic diversity, and species conservation |
The Video Recommended On Gene Mapping:
MCQs on Gene Mapping
Question: How are genetic maps used in the sequencing of whole genomes?
They provide a starting point for the sequencing process
They are used to determine the function of specific genes
They allow researchers to determine the exact location of mutations
They help to predict the phenotypic effects of genetic variants
Answer:Genetic maps provide a starting point for the sequencing of whole genomes by identifying the locations of genes on the chromosomes. This allows researchers to sequence specific regions of DNA and determine the order of genes along the chromosome.
Hence, the correct answer is option 1. They provide a starting point for the sequencing process.
Question: hat map unit (Centi Morgan) is adopted in the construction of genetic maps?
A unit of distance between two expressed genes, representing 10% cross-over.
A unit of distance between two expressed genes, representing 100% cross-over.
A unit of distance between genes on chromosomes, representing 1% cross-over
A unit of distance between genes on chromosomes, representing 50% cross-over.
Answer: The frequency of recombination between gene pairs on the same chromosome is used as an indicator of the distance between those genes, which can then be used to determine their relative positions in a genetic map. A map unit, or centimorgan (cM), is defined as the distance between genes for which a 1% crossover frequency occurs. This means there will be a 1% chance of an event of crossover between them during meiosis if two genes are one map unit apart. It therefore enables the understanding of gene linkage and inheritance patterns through this kind of mapping.
Hence the correct answer is Option (3) A unit of distance between genes on chromosomes, representing 1% cross-over.
Question: In the following questions, a statement of assertion (A) is followed by a statement of reason (R)
(1) If both Assertion & Reason are true and the reason is the correct explanation of the assertion, then mark A
(2) If both Assertion & Reason are true but the reason is not the correct explanation of the assertion, then mark B
(3) If Assertion is true statement but Reason is false, then mark C
(4) If both Assertion and Reason are false statements, then mark D
Assertion: Gene mapping can be used to study the evolution of species.
Reason: By comparing the location of genes between different species, researchers can gain insights into the evolutionary history of those species.
Both Assertion (A) and Reason (R) are the true and Reason (R) is a correct explanation of Assertion (A).
Both Assertion (A) and Reason (R) are the true but Reason (R) is not a correct explanation of Assertion (A).
Assertion (A) is true and Reason (R) is false.
Assertion (A) is false and Reason (R) is true.
Answer: Gene mapping can be used to study the evolution of species. By comparing the location of genes between different species, researchers can gain insights into the evolutionary history of those species. For example, researchers can use gene mapping to compare the location of genes between humans and other primates, shedding light on the evolutionary relationships between these species.
Hence, the correct answer is option 1 Both Assertion (A) and Reason (R) are true and Reason (R) is a correct explanation of Assertion (A).
Also Read:
Frequently Asked Questions (FAQs)
Gene mapping gives way to the identification of disease genes and prediction of disease risk, guides the process of genetic counselling, and enables personalized treatments free of adverse reactions by developing treatments tailor-made for every individual genetic profile.
Among those to be encountered are the challenges of the complexity of the structure of the genome, high-resolution techniques of mapping, and several ethical considerations for genetics information handling together with genomics data interpretation.
Gene mapping is the procedure through which the loci of genes are determined in chromosomes. It holds great importance for understanding heredity traits, inheritance of genes for disease transmission, and even population genetics which opens opportunities for medical research, agriculture, and evolutionary biology.
Linkage mapping describes the relative gene positions based on co-inheritance patterns of its genotypes in families or populations. In contrast, physical mapping determines the actual physical location of genes on the chromosome, most classically through DNA sequencing and cytogenetic techniques.
The techniques of gene mapping include linkage mapping using genetic markers like microsatellites and SNPs and physical mapping using methods such as restriction mapping and FISH. Modern sequencing technologies include whole-genome sequencing.