Gene to Protein - Transcription and Translation
The process from gene to protein is a fundamental concept in biology. It includes two main processes—transcription and translation. Genes store the instructions to make proteins in the living organism. The instructions are first copied into messenger RNA (mRNA) during transcription. This mRNA travels to ribosomes, and the protein synthesis starts with the genetic code in DNA. This flow of information is the core of gene regulation and gene expression.
This Story also Contains
- What is the Gene to Protein Process?
- Central Dogma: The Flow of Genetic Information
- Transcription And Translation
- The Role Of Gene Expression In Cellular Function
- Definition Of A Gene
- Types Of Gene
- Transcription: From DNA To mRNA
- Post-transcriptional Modifications In Eukaryotes
- Translation: From mRNA to Protein
- Stages Of Translation
- The Genetic Code
- Recommended video for Transcription and Translation
- MCQs on Central Dogma, Transcription, and Translation

The gene-to-protein process controls all life functions. Without transcription and translation, cells cannot survive. This flow of genetic information is the molecular basis of inheritance. It shows how DNA gives rise to proteins. It flows from DNA → RNA → Protein.
What is the Gene to Protein Process?
The gene-to-protein process makes proteins from genes. It starts with transcription and translation. In transcription, DNA makes mRNA. This mRNA carries the code to ribosomes. Then translation turns mRNA into proteins. These steps form protein synthesis. The gene-to-protein flow shows how traits form. It is the key to gene expression. Cells need genes to make proteins to survive.
Cellular conditions make a replica of the DNA molecule itself. This occurs just before the cell division, so that when the cell divides, each resulting cell will have a full complement of the genetic information of the parent cell.
It refers to the process by which a gene is expressed. It refers to the formation of an RNA molecule for a gene. It is essentially carried out by RNA polymerases, which are enzymes.
In other words, translation is the synthesis of a protein from the mRNA transcript. A ribosome reads the sequence that occurs via mRNA, uses nutrients with the assistance of tRNA, and then synthesises a corresponding amino acid series that forms a polypeptide chain.
Central Dogma: The Flow of Genetic Information
The central dogma of molecular biology explains how genetic information flows. It shows the path from DNA to RNA to protein. This idea was first given by Francis Crick in 1958. He explained how DNA makes RNA, then proteins. This flow is key to gene expression. Transcription and translation are part of this process. It connects the gene to the protein process in cells. The recent explanation of this key biological principle lies at the foundation of modern biology and explains the processes through which the genetic code contained within DNA is eventually transcribed into an active protein through an mRNA molecule.
Transcription And Translation
There are two major ways genes are expressed: transcription and translation. In essence, transcription is the process whereby a section of DNA gets copied into types of RNA—more specifically, mRNA. Copies of the mRNA then move the genetic information from the nucleus out into the cytoplasm to its counterpart, the ribosomes, for translation. For that reason, the ribosome will read the mRNA to synthesize a polypeptide chain for a functional protein in the process of translation.
The Role Of Gene Expression In Cellular Function
Gene expression is the process whereby information contained in DNA is transcribed into an active functional product, thereby considered as a protein or RNA molecule. This process is central to cellular function and organismal development. In this case, appropriate gene expression does take place in such a way that homeostasis within the cellular and organism levels is maintained. Diseases examined by cancer arise from the misexpression of genes.
Definition Of A Gene
It is defined as the linear sequence of nucleotides in DNA coding for the formation of certain proteins or an RNA molecule. It is the fundamental level of the inheritable unit found in chromosomes in the cell nucleus.
Structure Of The Gene Found On The Chromosome
Promoter: A DNA region where RNA polymerase binds to start the transcription.
Coding Sequence: Gene region from which RNA is transcribed, and later, portions where protein synthesis takes place.
Terminator: The nucleotide sequence signalling the end of transcription.
Types Of Gene
Protein-coding Genes: Information in these genes is for making proteins.
Non-coding Genes: It does not code for proteins, but can otherwise produce functional RNA molecules such as ribosomal RNA and transfer RNA.
Transcription: From DNA To mRNA
Transcription occurs in steps to make sure the conversion from DNA to RNA is precise. Each stage plays a vital role and controls how genes are expressed. It consists of three well-defined stages that include initiation, elongation, and termination:
Step 1: Initiation
The process of initiation is the beginning of RNA synthesis.
RNA polymerase binds to the promoter region of the DNA.
The transcription initiation complex is formed, and the process of unwinding od DNA double helix begins.
Step 2: Elongation
In elongation, RNA polymerase travels along the DNA to synthesize RNA.
RNA polymerase adds nucleotides to the growing mRNA strand in a 5' to 3' direction.
As RNA polymerase moves along the DNA, the double helix unwinds ahead of it and rewinds behind it.
Step 3: Termination
Termination signals the end of transcription and the release of the RNA molecule.
It is these specific terminator sequences in the DNA that cause the RNA polymerase to stop and release the mRNA transcript.
In eukaryotes, termination is followed by other processing steps.
Post-transcriptional Modifications In Eukaryotes
After transcription in eukaryotes, the RNA undergoes several changes to become functional mRNA. RNA processing helps in stability, proper export, and precise protein synthesis. Such processes are also important for gene regulation and expression. The post-transcriptional modifications are:
5' Capping
A transcription is initiated, and the 5' end of the mRNA is joined or capped with a modified guanine nucleotide that prevents the digestion of mRNA. It also has a significant role in ribosome binding to mRNA for translation.
Polyadenylation
The 3' end of mRNA is polyadenylated by the addition of a poly-A tail, which is believed to prevent degradation of the mRNA and also aids in the export of the mRNA from the nucleus.
Splicing
The non-coding regions (introns) are excised from the mRNA transcript and the coding regions (exons) are joined. This process is called RNA splicing, and a mature mRNA molecule is now ready for translation.
Translation: From mRNA to Protein
Translation is a precise process whereby the mRNA decoding pathway is used to translate a particular sequence of amino acids, which are induced to fold into a functional protein. Translation takes place in the cytoplasm with the help of types of RNA. The building blocks of proteins are amino acids that are linked in a specific order that is encoded by the base sequence of the mRNA in the process of translation.
mRNA (Messenger Ribonucleic Acid)
mRNA reads as the template for the synthesis of proteins. It translates the genetic information from DNA to the ribosome.
It is the in vivo specification of the amino acid sequence of polypeptide chains in the molecule of protein.
tRNA (Transfer RNA)
Molecules of this type carry amino acids to the ribosome and then learn the mRNA anticodon that would match the code which is encoded protein.
Ribosomes (rRNA and Protein Complex)
The ribosomes, composed of rRNA and proteins, carry out the process by which the information contained in the mRNA molecule is used to build the protein.
Stages Of Translation
The translation mechanism involves the ribosome reading the mRNA code. tRNA helps to bring the correct amino acid in the correct sequence. The process continues until a complete protein is formed. The details of the translation mechanism are listed below:
Initiation
Initiation occurs when the small ribosomal subunit attaches to the mRNA and a special initiator tRNA.
The small subunit is associated with the large subunit.
Elongation
During elongation, tRNAs bring amino acids to the ribosome in the order prescribed by the mRNA codons.
The actual polypeptide is elongated by peptide bonds that form between the amino acid residues.
Termination
Translation stops when the ribosome reaches a stop codon.
The ribosome releases the completed polypeptide, which will spontaneously fold into its final, functional conformation.
The Genetic Code
The genetic code is a set of rules whereby information encoded within the genetic material—in the form of DNA or RNA sequences—is translated into proteins by living cells. It is universal for almost all organisms and consists of nucleotide triplets called codons. Each codon corresponds to one amino acid or one of the signals to 'stop' while synthesizing the protein.
Codons and Anticodons
Triplets of nucleotides of mRNA that code for the amino acids. The sequence of three nucleotides on the mRNA, which codes for a particular amino acid. Anticodons are the triplets of nucleotides on the tRNA molecules that are complementary to codons.
Initiator And Terminator Codons
Translation initiates with an initiator codon (AUG) and terminates with a terminator codon (UAA, UAG, or UGA).
Redundancy and Universality of the Genetic Code
The genetic code is degenerate in the sense that more than one triplet code can code for one amino acid. It is, however, universal for almost all organisms.
Recommended video for Transcription and Translation
MCQs on Central Dogma, Transcription, and Translation
Question: Which of the following most accurately describes the central dogma of biology?
RNA to protein to DNA
DNA to RNA to protein
DNA to protein to RNA
Protein to RNA to DNA
Answer: The central dogma of biology dictates that the coded genetic information stored in DNA is transcribed into single-stranded RNA, which is then translated into protein.
The central dogma states that the pattern of information that occurs most frequently in our cells is:
From existing DNA to make new DNA (DNA replication)
From DNA to make new RNA (transcription)
From RNA to make new proteins (translation).
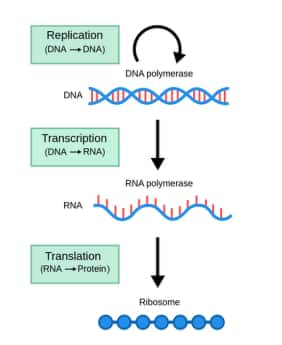
Hence, the correct answer is option 2) DNA to RNA to protein
Question: A transcription unit in DNA is defined primarily by the three regions in DNA and these are for upstream and downstream ends:
Repressor, Operator gene, Structural gene
Structural gene, Transposons, Operator gene
Inducer, Repressor, Structural gene
Promotor, Structural gene, Terminator
Answer: The unit is a distinct DNA segment transcribed into RNA. It is composed of three main regions, each with a unique role in the transcription process:
1. Promoter Region:
Position: Before the gene on the 5’ end of the DNA template.
Role: It is the RNA polymerase and transcription factor attachment site.
Characteristics: It features conserved sequences like the eukaryotic TATA box or prokaryotic Pribnow box, and may include upstream elements affecting transcription.
2. Structural Gene:
Position: After the promoter.
Role: Contains the actual DNA sequence that gets transcribed into RNA types such as mRNA, tRNA, and rRNA.
Key Aspect: In eukaryotes, it includes exons (coding regions) and introns (non-coding regions). The RNA polymerase synthesizes RNA complementary to the DNA's 3’ → 5’ direction.
3. Terminator Region:
Position: Beyond the structural gene on the 3’ end of the DNA template.
Role: Indicates transcription's conclusion and causes RNA polymerase release from DNA.
Characteristics: These can be defined by specific RNA sequences or structures like hairpin loops in prokaryotes.
Hence, the correct answer is option 4) Promotor, Structural gene, Terminator.
Question: In the process of translation, which of the following is the translational site of mRNA?
Nucleus
Nucleolus
Golgi body
Ribosomes
Answer:
The translation process uses the ribosome as the decoding site for mRNA. The elongation phase of protein synthesis continues in this step as complexes of amino acids bonded to tRNA attach to the codons on the mRNA in a step-by-step sequence. This happens due to the complementary base pairing between the tRNA anticodon and the codon on mRNA. With the movement of the ribosome along mRNA, amino acids start getting added in a sequence. These sequences of polypeptides are determined by DNA and written into mRNA.
Hence, the correct answer is Option 4) ribosomes.
Also Read:
Frequently Asked Questions (FAQs)
The former occurs in the cytoplasm of prokaryotes, and those processes are not as complex as eukaryotic ones while the mechanisms controlling them differ.
Mutations alter the DNA sequence; hence, they cause changes in the processes of transcription and translation, which might lead to an occurrence of sickness.
Transcription is the process in which a copy of the gene's DNA sequence is made into a complementary mRNA; translation is the process by which mRNA is translated into a protein.
The steps involved are initiation, elongation, and termination, with post-transcriptional modifications taking place in eukaryotes.
The genetic code consists of the codons of the mRNA, which specify the amino acids of the protein. It is universal and redundant.