Process of Translation in Biology: Definition, Steps and Process
Translation is a crucial step in the process of protein synthesis, where the genetic information carried by mRNA is decoded to make proteins. The proteins are necessary for various cellular functions and are formed by joining amino acids in the correct sequence. The ribosomes read mRNA, and tRNA brings amino acids into the polypeptide chain. This step is part of the molecular basis of inheritance, as it directly expresses the information stored in DNA.
This Story also Contains
- Definition Of Translation
- Key Components Of Translation
- Stages Of Translation
- Translation Mechanism Details
- Post-Translational Modifications
- Regulation of Translation
- Common Mistakes In Translation
- Applications Of Translation Study
- Recommended video for the Process Of Translation
- MCQs on Translation

Translation is one of the major steps in central dogma of molecular biology which describes the flow of genetic information from DNA to RNA to protein. It is important to understand this process as it explains how genetic instructions are carried out in organisms. It also highlights the role of RNA and ribosomes in decoding genetic code into functional proteins.
Definition Of Translation
Translation is one of the major biological processes by which the ribosomes make proteins using mRNA as a template. The process is critical in gene expression and regulation that allows genetic information in DNA to be converted into active proteins. These proteins play crucial roles in nearly all cellular functions.
Translation plays a crucial role in gene expression and regulation, as it decides both the type and amount of proteins. It is rather highly regulated that the wrongly synthesised proteins are mostly not read. The errors in translation produce malfunctioning proteins that can cause diseases and developmental problems.
Diagram: Process Of Translation
The diagram illustrates how proteins are made using the genetic code carried by mRNA. It shows the binding of mRNA to the ribosome and how it reads codon. Each codon is paired with a specific amino acid carried by the tRNA. These amino acids are linked together to form a polypeptide chain.
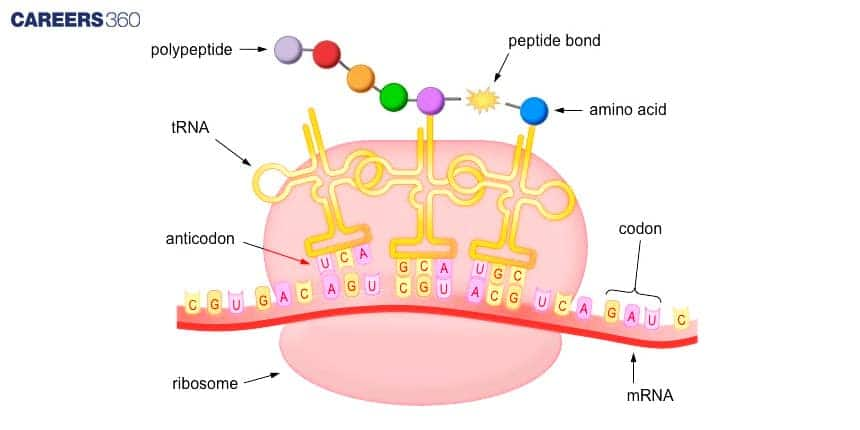
Key Components Of Translation
The process of translation involves several elements working together to ensure accurate protein synthesis. The components help decode the mRNA into sequences of amnio acids. The key components of translation are as follows:
Messenger RNA (mRNA)
mRNA is a single positively charged molecule that helps carry information from DNA at the top to the ribosomes.
It has a chain of nucleotides that holds information on the sequence of amino acids for a certain protein throughout the cellular environment.
The structure undergoes RNA processing and contains a 5' cap and a 3' poly-A tail, which both protect the mRNA from degradation.
Ribosomes
Ribosomes are protein manufacturing machines. A ribosome has two subunits, the small and large subunits.
The small and large subunits in prokaryotes measure 30S and 50S respectively, while in eukaryotic cells they measure 40S and 60S respectively.
In other words, ribosomes mediate messenger RNA transfer RNA binding and catalyze peptide bonds between amino acids.
Transfer RNA (tRNA)
The tRNA molecule acts as an adapter that picks up and carries the appropriate amino acid corresponding to the particular codon in the mRNA strand.
Every tRNA has some region for the anticodon and a site for attaching a particular amino acid.
Correct pairing of anticodons with mRNA codons guarantees that amino acids will be added to the polypeptide chain in the correct order by protein synthesis.
Amino Acids
The building blocks of the proteins are amino acids. There are 20 different amino acids. Each one has a different side chain.
It determines the structure and thus function of a protein which is how the nature and sequence of the amino acid in the chain.
During translation, these are linked by peptide bonds to form a polypeptide chain.
Enzymes And Factors
Many enzymes and factors are involved in ensuring translation occurs with appropriate accuracy and speed.
Initiation factors help bind the ribosomal subunits and the first tRNA to the mRNA strand.
Elongation factors assist in adding more amino acids to the polypeptide chain, while termination factors help in releasing the completed polypeptide from the ribosome once the stop codon has been encountered.
Stages Of Translation
Translation occurs in three main stages and each plays a vital role in building protein from the mRNA template. The process starts with binding of mRNA to ribosomes and ends when the protein chain is formed and released. The stages of translation are mentioned below:
Initiation
The first step to take is the initiation step by the process of translation. This is when the translational apparatus gets assembled on the mRNA molecule.
In a simple sense, this occurs with the binding of the small ribosomal subunit with mRNA at its initiation codon, which is AUG.
These are assisted in doing this by initiation factors. Initiator t-RNA, charged with methionine, base pairs with the initiation codon and then the large ribosomal subunit joins the complex thus forming an initiation complex.
Elongation
In the elongation stage, amino acids are added to the growing polypeptide chain one by one.
Elongation is a three-step process: codon recognition, formation of peptide bonds, and translocation.
First, the appropriate tRNA diffuses into the A site of the ribosome so that its anticodon bonds with the mRNA codon.
Then, a peptide bond is formed between the amino acid attached at the P site of the tRNA and the amino acid in the A site.
Translocation
The final stage of each elongation cycle is translocation.
The ribosome moves one codon along the mRNA–shifting the tRNAs and the associated growing amino acid chain.
The tRNA in the P site translocates to the E site and is released, and the tRNA in the A site, now holding the growing chain of polypeptides, translocates to the P site.
In this manner, this cycle (initiation, elongation, translocation) is repeated according to the sequence of nucleotides in the mRNA – adding amino acids one at a time.
Termination
Termination also occurs when a stop codon pattern is reached on the mRNA. No tRNA anticodon corresponds to stop codons.
Instead, the RF release factor binds the stop codon and induces the disassembly of the translation complex.
The newly synthesised polypeptide is released from the ribosome, and the two subunits of the ribosome dissociate and can start another round of translation.
Translation Mechanism Details
The translation mechanism involves the ribosome reading the mRNA code. tRNA helps to bring the correct amino acid in the correct sequence. The process continues until a complete protein is formed. The details of translation mechanism are listed below:
Initiation
Initiation follows some important steps. The small ribosomal subunit joins to the mRNA close to the start codon.
The initiator tRNA with methionine is placed over the start codon—AUG—by the initiation factors at the AUG codon.
The big ribosomal subunit is added lastly to the complex. One entire initiation complex is made. Translation starts instantly.
Elongation
Elongation occurs by a cyclic method of similar repeated steps.
The ribosome promotes the correct matching of tRNA anticodons with mRNA codons in the A site.
A peptide bond is then formed between an amino acid that is in the P site and an amino acid that is in the A site by the peptidyl transferase activity of the ribosome.
Then the ribosome translocates, shifting the tRNA to the P site, itself moving one codon upstream on the mRNA; then a new tRNA enters the A site to initiate the process anew.
Termination
When a stop codon reaches the A site, release factors bind the ribosome and implement polypeptide chain release.
Then the ribosome disassembles into its subunits; mRNA and tRNA are released and translation is complete.
The newly synthesised polypeptide folds. Possibly further post-translations take place.
Post-Translational Modifications
Consequently, post-translational modifications become obligatory for the final functional forms of proteins. PTMs are capable of modulating the activity, stability, and interactions of any protein. Examples of common PTMs include phosphorylation, where a phosphate group is added and this turns on or off the activity, glycosylation includes addition of sugar moieties to proteins, and proteolytic cleavage, meaning certain peptide linkages have to be cleaved to turn on or off the function of proteins.
Regulation of Translation
The controlled translation allows the correct number of proteins to be produced. This is also the site where both microRNAs and RNA interference bind with the mRNA preventing translation. Global regulation in response to nutrient availability will occur as well so the cells can regulate another environmental factor.
Common Mistakes In Translation
Different types of mutations can occur by translation errors include missense mutations, nonsense mutations, and frameshift mutations. Missense mutations result from misinserted amino acids, leading to changes in protein activities. Nonsense mutations provide a premature stop codon, which results in truncation of the peas. An example of frameshift mutations includes insertions or deletions, resulting in differing reading frames and thus different downstream amino acids.
Applications Of Translation Study
Understanding how translation takes place has much wider applications in biology and medicine. For example, new antibiotics can be designed which would specifically target the bacterial ribosomes, drugs that would help in the treatment of human genetic disorders, and current advances in biotechnology where genes and proteins are synthetically designed for use in industry and therapy.
Recommended video for the Process Of Translation
MCQs on Translation
Q1. In the process of translation, which of the following is the translational site of mRNA?
Nucleus
Nucleolus
Golgi body
Ribosomes
Correct answer: 4) ribosomes.
Explanation:
The translation process uses the ribosome as the decoding site for mRNA. The elongation phase of protein synthesis continues in this step as complexes of amino acids bonded to tRNA attach to the codons on the mRNA in a step-by-step sequence. This happens due to the complementary base pairing between the tRNA anticodon and the codon on mRNA. With the movement of the ribosome along mRNA, amino acids start getting added in a sequence. These sequences of polypeptides are determined by DNA and written into mRNA.
Hence the correct answer is Option 4) ribosomes.
Q2. SD region in prokaryotic mRNA is a part of ______.
TATA BOX
RBS (Ribosome Binding Site)
ASD region
Promoter
Correct answer: 2) RBS (Ribosome Binding Site)
Explanation:
The Shine-Dalgarno (SD) region is a ribosomal binding site in prokaryotic messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA. The Shine-Dalgarno (SD) region is a ribosomal binding site in prokaryotic messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA, ensuring proper translation initiation. The sequence is complementary to a region of the 16S rRNA of the small ribosomal subunit, facilitating the alignment of the ribosome with the mRNA to begin protein synthesis.
Hence, the correct answer is option 2) RBS (Ribosome Binding Site)
Q3. Select the correct option w.r.t process of translation in prokaryotes
i. During the initial phase, GTP acts as a source of energy.
ii. There are three initiation factors (IF 1, IF 2 and IF 3) that help the assembly of the initiation complex.
iii. The first mRNA is translated by formyl aminoacyl tRNA.
All of the above
i & iii
i & ii
None of the above
Correct answer: 1) All of the above
Explanation:
Translation also occurs in three stages: initiation, elongation, and termination. During the initiation stage of translation in prokaryotes, several key components assemble to form the translation complex. These components include the small and large ribosomal subunits, the mRNA that will be translated, the first (formyl) aminoacyl tRNA (charged with the first amino acid), GTP (which provides energy for the process), and three initiation factors (IF1, IF2, and IF3) that facilitate the assembly of the initiation complex. The small ribosomal subunit binds to the mRNA, and the first aminoacyl tRNA pairs with the start codon on the mRNA. These steps set the stage for the elongation phase, where the protein is synthesized.
Hence, the correct answer is option 1) All of the above.
Also Read:
Frequently Asked Questions (FAQs)
Translation is the synthesis of proteins from mRNA, while translocation is the movement of the ribosome along the mRNA during translation.
tRNA carries specific amino acids to the ribosome and matches them to mRNA codons using its anticodon.
The three steps are initiation, elongation, and termination.
An anticodon is a three-nucleotide sequence on tRNA that pairs with the complementary codon on mRNA..