Difference Between Prokaryotic and Eukaryotic Replication
DNA Replication in a cell is a process of copying genetic material before cell division. The difference between DNA Replication in prokaryotes and eukaryotes is mainly in location of occurrence, complexity, and speed of replication. Prokaryotic replication happens in circular chromosomes with a single origin. Eukaryotic replication has multiple origins on linear chromosomes. The difference between DNA replication in prokaryotes and eukaryotes is part of the molecular basis of inheritance, showing how DNA is copied in different cells.
This Story also Contains
- What Is DNA Replication?
- DNA and Its Components
- Essential Components Involved in Replication
- DNA Replication in Prokaryotes
- DNA Replication in Eukaryotes
- Differences between Prokaryotic and Eukaryotic Replication
- Similarities between Prokaryotic and Eukaryotic Replication
- MCQs on Prokaryotic and Eukaryotic Replication

Eukaryotic replication is more complex and slower, and it needs additional enzymes and proteins. This article includes replication of DNA biology, the structure and components of DNA, essential components involved in replication, and the differences and similarities between prokaryotic and eukaryotic replication. Understanding the difference between DNA replication in prokaryotes and eukaryotes is important for genetics and molecular biology.
What Is DNA Replication?
DNA replication refers to a process in which a cell makes an exact copy of its DNA. It provides each new cell with the full set of genetic material. Every daughter cell at the time of cell division gets an identical copy. This is a process that is not only important for the genetic continuity of living organisms. It is also necessary for growth, development, and other functions by providing the system with identical copies of the required set of genetic information in succeeding generations.
DNA and Its Components
DNA stands for deoxyribonucleic acid. It is a double helix structure composed of two strands. The building blocks of DNA are nucleotides. Each nucleotide consists of a phosphate group, a deoxyribose sugar, and nitrogenous bases. These nitrogenous bases are adenine, thymine, cytosine, and guanine. Adenine always pairs with thymine, and likewise, cytosine always pairs with guanine. These base pairs form the steps of the DNA ladder.
Essential Components Involved in Replication
DNA replication is a complex process that needs the help of many proteins. There are different enzymes used in DNA replication. These components work together to ensure the DNA is copied correctly and quickly.
DNA Polymerase: This is an enzyme that synthesises new strands of DNA by adding nucleotides that complement the template strand.
Helicase: An enzyme breaking the DNA double helix at the replication fork.
Primase: An enzyme synthesising RNA primers necessary to initiate DNA synthesis.
Ligase: This is also an enzyme. It joins Okazaki fragments on the lagging strand.
Single-Strand Binding Proteins (SSBs): They stabilise the unwound DNA strands, preventing them from re-annealing.
Sliding Clamps: Keep DNA polymerase bound to the template strand.
DNA Replication in Prokaryotes
DNA replication in prokaryotes is a quick and efficient process. It takes place in the cytoplasm and begins at a single point on the circular DNA. This point is called the origin of replication (OriC). The process includes four main steps: initiation, elongation, termination, and proofreading. Prokaryotic DNA replication uses several enzymes that ensure accuracy and speed. The replication mechanism in prokaryotic cells is described below:
Origin of Replication (OriC): Replication starts at a single, unique site on the circular DNA molecule known as the origin of replication.
Initiation: Helicase opens the DNA. The primase lays down an RNA primer.
Elongation: In this process, a new DNA strand is being synthesised on the lagging strand in a 5' to 3' direction; this is done by DNA polymerase III.
Termination: The final process, wherein the replication forks finally meet at the termination site.
Replication Fork and Enzyme Functions
DNA Polymerase III: main enzyme for DNA synthesis.
Helicase: unwinds the DNA helix. Primase: creates RNA primers for initiation.
DNA Polymerase I: Replaces RNA primers with DNA.
Ligase: Seals gaps between DNA fragments.
Replication Speed and Accuracy
Replication in prokaryotes is very fast, about 1000 nucleotides per second.
The process is highly accurate due to the proofreading function of DNA polymerase.
Repair enzymes help maintain the quality of the replicated DNA.
DNA Replication in Eukaryotes
DNA replication in eukaryotes is more complex than in prokaryotes. It takes place in the nucleus and involves larger linear chromosomes. To copy the entire genome efficiently, eukaryotic DNA replication begins at multiple origins of replication. The process includes initiation, elongation, and termination, supported by several specialized enzymes. The replication process in eukaryotic cells is described below:
Multiple Origins of Replication: Eukaryotic chromosomes have multiple origins so that the replication of a larger genome can be completed in time.
Initiation: The origin recognition complex binds to the origins and helicase unwinds the DNA.
Elongation: The chains are initiated by DNA polymerase α. It is further extended by DNA polymerases δ and ε.
Termination: It ends when the replication forks meet or at the telomeres in linear chromosomes.
Replication Fork and Enzyme Functions
DNA Polymerase: Initiates DNA synthesis by extending RNA primers.
DNA Polymerase δ and ε: Extend leading and lagging strands, respectively.
Helicase: Unwinds DNA.
Ligase: Joins Okazaki fragments.
Telomerase: Extends telomeres to avoid chromosome shortening.
Replication Speed and Accuracy
Slower than in prokaryotes, with about 50 nucleotides per second on average.
Very accurate, owing to the inbuilt complex proofreading and repairing mechanisms.
These mechanisms ensure faithful copying of genetic material for cell division.
Differences between Prokaryotic and Eukaryotic Replication
This is an important topic in biology and genetics. Understanding the differences in DNA replication between prokaryotes and eukaryotes helps us learn how cells copy their DNA with accuracy and speed. It is one of the important differences and comparison articles in biology. The differences are given below-
Feature | Prokaryotic Replication | Eukaryotic Replication |
Location | ||
Origin of Replication | Single (OriC) | Multiple origins per chromosome |
DNA Polymerases | DNA polymerase III for elongation | DNA polymerase α, δ, ε |
Speed | Faster (≈1000 nucleotides/second) | Slower (≈50 nucleotides/second) |
Genomic Structure | Circular DNA | Linear chromosomes with telomeres |
Initiation Complex | Simple | Complex (involving ORC, helicase loading proteins) |
Number of Replication Forks | Typically 2 (from single origin) | Multiple forks due to multiple origins |
Okazaki Fragment Size | Longer fragments | Shorter fragments |
Telomere Handling | Not applicable | Telomerase extends telomeres |
Proofreading Mechanisms | Present, primarily by DNA polymerase III | Extensive, involving multiple polymerases and repair pathways |
Replication Timing | Continuous | S-phase specific |
Similarities between Prokaryotic and Eukaryotic Replication
DNA replication in prokaryotes and eukaryotes has many similarities. Both follow the same basic steps. The process starts by unwinding the DNA. The same types of enzymes are used. Replication moves in both directions. These similarities in DNA replication help keep genetic information safe. It is a key part of the central dogma of molecular biology, where DNA is copied before being used to make RNA and proteins.
Fundamental Process: It consists of the unwinding of the DNA helix, synthesis of primers, chain elongation by DNA polymerase, and ligation of fragments.
Helicase, primase, DNA polymerase, and ligase are used in both.
Bidirectional Replication: Replication occurs in both directions from the origin in both prokaryotes and eukaryotes.
Proofreading and Repair: It has mechanisms for replication fidelity, along with error checking/correction methods for correct replication.
MCQs on Prokaryotic and Eukaryotic Replication
Question: The correct order of enzymes involved in DNA replication of prokaryotes
(1) Helicase - Single strand DNA binding protein - Topoisomerase - Primase - DNA Pol III -DNA Pol I - Ligase
(2) Topoisomerase - Helicase - Single strand DNA binding protein - Primase - DNA Pol III -DNA Pol I - Ligase
(3) Primase - DNA Pol I - DNA Pol II - DNA Pol III - Ligase
(4) Ligase - Primase - DNA Pol I - DNA Pol II - DNA Pol III - Topoisomerase
Answer: Prokaryotic replication starts with DNA unwinding by the helicase enzyme. SSB proteins stabilise the strands from rewinding. Supercoiling is controlled by topoisomerase. Primase synthesises the starting material primers which are extended by DNA Pol III and finally removed by Pol I and The Ligase seals the gaps. This is the correct order of the enzymes in DNA replication in Prokaryotes. Hence option A is correct.
Topoisomerase prevents supercoiling and it is not the first enzyme involved in replication. Thus option B is incorrect.
Primase can synthesise the primers and initiate replication only if the DNA strands are separated. Thus option C is incorrect.
Ligase is the sealing enzyme the last enzyme involved in the DNA replication process. Hence option D is incorrect.
Hence, the correct answer is Option (1) Helicase - Single strand DNA binding protein - Topoisomerase - Primase - DNA Pol III -DNA Pol I - Ligase.
Question: Assertion: DNA replication in prokaryotes starts from a single origin of replication.
Reason: Prokaryotic genomes are circular in nature.
Both Assertion and Reason are true and Reason is the correct explanation for Assertion.
Both Assertion and Reason are true but Reason is not the correct explanation for Assertion.
Assertion is true but Reason is false.
Assertion is false but Reason is true.
Answer: In prokaryotes, DNA replication starts from a single origin of replication. This is because the prokaryotic genome is circular. The replication starts at a specific point on the circular genome and proceeds in two directions, eventually meeting on the opposite side of the circular genome.
The circular nature of the prokaryotic genome necessitates the presence of a single origin of replication because replication has to start at a specific point and proceed in both directions until it meets on the opposite side. Therefore, both the assertion and reason are true, and the reason correctly explains the assertion.
Hence, the correct answer is Option 1) Both Assertion and Reason are true and Reason is the correct explanation for Assertion.
Question: The mechanism of eukaryotic DNA replication differs from prokaryotic DNA replication due to
Use of DNA primer rather than RNA primer.
Different enzymes for synthesis of lagging and leading strands.
Discontinuous replication occurs rather than semi-discontinuous replication.
Unidirectional replication rather than semi-discontinuous replication.
Answer: DNA Replication in Eukaryotes -
In eukaryotes, one strand of DNA is synthesized continuously but the other one is synthesized in a semi-discontinuous manner and made of Okazaki fragments.
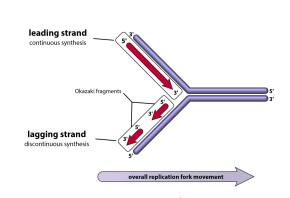
Hence, the correct answer is option 3)Discontinuous replication occurs rather than semi-discontinuous replication.
Also Read-
Frequently Asked Questions (FAQs)
The replication in eukaryotic cells is more complex, making it necessary to have multiple origins to complete the process on time.
Telomeres are discrete, non-coding, repetitive sequences located at the linear chromosomes' terminal. They save the ends of linear chromosomes from degradation and prevent them from being recognised as damaged DNA.
While proofreading is an intrinsic property of DNA polymerases in all living organisms, both prokaryotes and eukaryotes, the latter have additional repair pathways brought about by the complexity of their genomes.
The chief difference is in prokaryotic replication, which prudently contains one origin of replication, while eukaryotic replication contains multiple origins.
Basically, prokaryotic replication is much faster since it contains less regulatory apparatus and its replication machines are simple in design.
