Replication of DNA: Know Process and Diagram
The replication of DNA is the process by which a cell makes copies of its DNA. This process occurs in all living organisms. It is an important part of the biological inheritance, cell division during growth and repair. The process also ensures that each cell receives its own copy of the DNA.
This Story also Contains
- What is DNA Replication?
- DNA Structure
- Enzymes involved in DNA Replication
- Steps of DNA Replication
- Semi-Conservative Nature of DNA Replication
- DNA Replication in Prokaryotes vs Eukaryotes
- Recommended Video for Replication of DNA
- MCQs on Replication of DNA

DNA is a double helical structure with two complementary strands. The strands run opposite to each other. During replication, these strands get separated, and each strand of the original DNA serves as a template for the production of the new strand. It is a semi-conservative mode of DNA replication. DNA Replication is a topic of the chapter Molecular Basis of Inheritance. It is an important chapter in the Biology subject.
What is DNA Replication?
DNA replication is a biological process by which a cell makes a duplicate copy of its DNA, essentially making two copies out of a single DNA molecule. This step is accomplished before cell division to ensure that the new cell produced at the end of the process has an exact, complete set of genetic material. Hence, DNA replication is the basis for life processes such as growth, development, and reproduction.
DNA Structure
The structure of the DNA molecule was elucidated by James Watson and Francis Crick in 1953. It was described as being a double helix very similar in structure to a twisted ladder. The backbone of this ladder was represented to be composed of a sugar-phosphate backbone, with the rungs represented by the nitrogenous base pairs: adenine with thymine (A-T) and cytosine with guanine (C-G).
The DNA strands are anti-parallel; they run in opposite directions. One runs 5' to 3', and another runs 3' to 5'. This orientation is important for replication, for the DNA polymerases can add nucleotides to the 3' end of the growing DNA strand only.
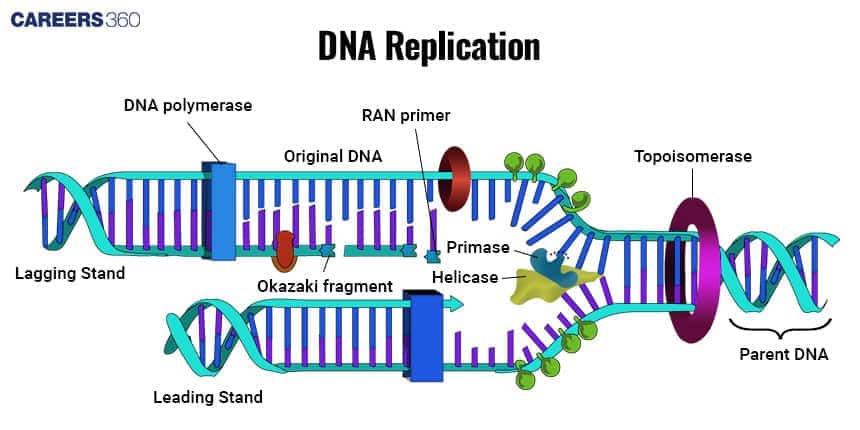
Enzymes involved in DNA Replication
DNA replication is facilitated and accurately done simultaneously due to the various roles played by different enzymes. These enzymes used in DNA replication:
Helicase: These open the double helix of DNA into two single-stranded templates.
Primase: These enzymes synthesise short RNA primers required for DNA polymerases to initiate nucleotide incorporation.
DNA Polymerase: Adds nucleotides to the growing DNA strand in a 5' to 3' direction. In prokaryotic cells, it is mainly DNA Polymerase III that synthesises the new DNA, while replacing RNA primers with DNA is done by DNA Polymerase
Ligase: Seals nicks and joins Okazaki fragments on the lagging strand to provide a continuous DNA strand.
Topoisomerase: Prevents the DNA from supercoiling and tangling ahead of the replication fork.
Single-Strand Binding Proteins (SSBs): Bind unwound DNA strands to prevent them from reannealing.
Steps of DNA Replication
DNA Replication is the process by which one strand of the parent DNA acts as a template to produce a complementary daughter strand. The mechanism of DNA replication consists of three main stages: initiation, elongation, and termination.
Initiation
Replication begins at specific regions of the DNA molecule known as the origins of replication. In these regions, the helix is unwound initially by helicase to generate a replication fork. The binding of single-strand binding proteins would stabilise the unwound DNA while RNA primers are made by a primase, which acts as a starting point for actual DNA polymerases.
Elongation
During the process of elongation, DNA polymerase III adds nucleotides to synthesise the new DNA strands. The leading strand is continuously synthesised toward the replication fork while the lagging strand is synthesised in short segments called Okazaki fragments. As described earlier, these RNA primers are later replaced with DNA by DNA polymerase I and the Okazaki fragments are joined into a continuous strand by DNA ligase.
Termination
Replication is completed when the entire DNA molecule has been duplicated. In eukaryotic cells, the ends of linear chromosomes are capped by telomeres, which also prevent the loss of genetic material during replication. Telomerase, an enzyme, lengthens the end telomeres and enables the complete replication of chromosome ends.
Diagram: DNA Replication
The diagram below shows the process of replication in DNA.
Semi-Conservative Nature of DNA Replication
In the semi-conservative model for DNA replication put forward by Watson and Crick, each of the new DNA molecules contains one old strand and one newly synthesised strand. Meselson and Stahl Experiment supported this model by showing, after a single round of replication, that the DNA consisted of one old and one new strand associated with it, fitting the mechanism of replication as being semi-conservative.
DNA Replication in Prokaryotes vs Eukaryotes
The replication process in prokaryotes and eukaryotes is quite different in terms of origin and the number of proteins involved. The replication process of prokaryotes and eukaryotes is mentioned below:
DNA Replication in Prokaryotes | DNA Replication in Eukaryotes |
Single origin of replication | Multiple origins of replication |
Faster replication due to a simpler structure | Slower replication due to complex chromatin structure |
Circular DNA | Linear DNA with telomeres |
Fewer proteins involved | More controlling elements and proteins involved in the process |
Recommended Video for Replication of DNA
MCQs on Replication of DNA
Q1. Semi-conservative replication of DNA was first demonstrated in:
Option 1: Escherichia coli
Option 2: Streptococcus pneumoniae
Option 3: Salmonella typhimurium
Option 4: Drosophila melanogaster
Correct answer: 1) Escherichia coli
Explanation:
The semi-conservative replication of DNA was initially demonstrated by scientists Meselson and Stahl in the year 1958. They performed a significant experiment that offered substantial proof in favour of this specific DNA replication model. Here's a succinct summary of their research:
Key Experiment: Meselson-Stahl Experiment
Aim: To evaluate the hypothesis on DNA replication, whether it's conservative, semi-conservative, or dispersive.
Procedure:
Isotopic Marking:
Employed nitrogen isotopes to differentiate old and new DNA strands.
Cultivated E. coli in a medium with heavy nitrogen (^15N), which got incorporated into DNA.
Subsequently, the bacteria to a medium with light nitrogen (^14N).
Centrifugation Process:
Isolated DNA post one and two replication cycles, then utilized density gradient centrifugation in caesium chloride.
Observations:
After one round of duplication: Noticed a single band of intermediate density, indicating DNA with one ^15N and one ^14N strand.
After two rounds of duplication: Identified two bands, one with intermediate density (^15N-^14N hybrid) and another with light density (^14N only).
Hence, the correct answer is option 1) Escherichia coli
Q2. The experimental proof for semiconservative replication of DNA was first shown in a
Option 1: Plant
Option 2: Bacterium
Option 3: Fungus
Option 4: Virus
Correct answer: 2) Bacterium.
Explanation:
Observations of experiments done by Meselsons and Stahl -
The DNA that was extracted from the culture of E.coli bacteria one generation after the transfer from 15 N to 14 N medium had a hybrid or intermediate density or DNA extracted another generation (40 minutes) was composed of an equal amount of this hybrid DNA
Semi-conservative DNA replication was first shown in Bacterium Escherichia coli by Matthew Meselson and Franklin Stahl.
Hence, the correct answer is option 2)Bacterium.
Q3. Assertion (A): The role of the Origin of Replication (ori) in any plasmids to initiate the replication
Reason (R): Initiation of replication is the beginning of replication on chromosomes, plasmids, or viruses.
Option 1: Both A and R are true and R is the correct explanation of A.
Option 2: Both A and R are true but R is not the correct explanation of A.
Option 3: A is true but R is false.
Option 4: A is false but R is true.
Correct answer: (2) Both A and R are true but R is not the correct explanation of A.
Explanation:
Initiation of replication is the beginning of replication on chromosomes, plasmids, or viruses. For short DNAs, such as bacterial plasmids and small viruses, a single-origin works quite well. A certain segment in a genome is where replication starts, commonly referred to as the replication origin.
Hence, the correct answer is Option (2) Both A and R are true but R is not the correct explanation of A.
Also Read-
Frequently Asked Questions (FAQs)
DNA polymerase proofreads and edits its work during synthesis, and the remaining errors are repaired by the mismatch repair mechanisms.
So that at the time of cell division, every new cell formed must get an identical copy of the DNA.
Whereas prokaryotes have one origin of replication and circular DNA, eukaryotes have multiple origins of replication and linear DNA with telomeres.
The main ones are the helicase, primase, DNA polymerase, ligase, topoisomerase, and single-strand binding proteins involved in the unwinding, synthesis, and stabilisation of DNA.
The semi-conservative model means that each new DNA molecule contains one old strand and one newly made strand.
