Recombinant DNA Technology: Definition, Tools, Steps, Examples, Applications & Diagram
Recombinant DNA technology is one of the key foundations of modern biotechnology and has utterly transformed medicine, agriculture, and industrial processes. It is the technology of joining DNA molecules from two species to yield a recombinant with a useful scientific investigation or practical application.
- Definition of Recombinant DNA Technology
- What is Recombinant DNA Technology?
- Key Principles and Mechanisms
- Tools of Recombinant DNA Technology
- Recommended video on Recombinant DNA Technology
- Steps in Recombinant DNA Technology
- Amplification of Gene of Interest
- Inserting the Gene of Interest into a Vector
- Introduction of Recombinant DNA into Host Cells
- Obtaining the Foreign Gene Product
- Applications of Recombinant DNA Technology
- DNA Cloning

Definition of Recombinant DNA Technology
Recombinant DNA technology is the set of methods used for joining together DNA segments from two or more different sources. Recombinant DNA molecules are inserted into an organism to produce new genetic combinations that lead to practical applications in medicine, agriculture, and biotechnology.
The technology of recombinant DNA was founded in the 1970s by efforts through the laboratories of such pioneers as Paul Berg, Herbert Boyer, and Stanley Cohen. These seminal experiments proved that DNA from different sources can be spliced together and replicated in bacterial cells, a key basic principle of genetic engineering.
Recombinant DNA technology is very important in several scientific and industrial sectors. It allows for the production of valuable pharmaceuticals, the development of genetically modified organisms for better agriculture, and enzymes for industrial processes. Equally important is its ability to help push medical research and treatment in new directions.
What is Recombinant DNA Technology?
Recombinant DNA technology is the general technique used to cut, join, and clone DNA fragments into host organisms. To alter the DNA of an organism by putting new genetic material into its cell, the source of this new genetic material has to be recombined DNA from two different sources, and then the recombinant DNA is put into the recipient host cell to reproduce and express.
Key Principles and Mechanisms
The normal steps involved in recombinant DNA technology include isolation of genetic material, restriction enzyme digestion, amplification of the gene of interest, ligation into the vector, transformation or transfection to the recipient, and expression of the recombinant gene. All these steps involve very specific tools and techniques to realize a particular genetically modified state.
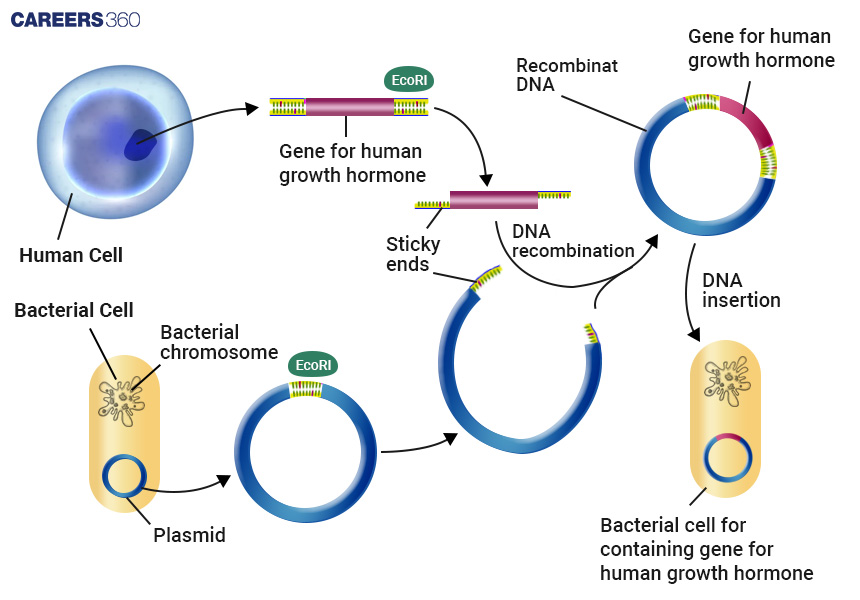
Diagram: Overview of the Recombinant DNA Process
Tools of Recombinant DNA Technology
Table: Key Tools and Their Functions in Recombinant DNA Technology
Tool | Functions | Example |
Restriction Enzymes | Cut DNA at specific sequences | EcoRI, HindIII |
DNA Ligase | Joins DNA fragments together | T4 DNA Ligase |
Vectors | Carry foreign DNA into host cells | Plasmids (pBR322), Bacteriophages |
Host Cells | Replicate and express recombinant DNA | E. coli, Yeast |
Polymerase Chain Reaction (PCR) | Amplifies DNA fragments | Taq Polymerase |
Recommended video on Recombinant DNA Technology
Restriction Enzymes
Restriction enzymes are molecular scissors, cleaving DNA at specific sequences known as restriction sites. Different enzymes recognize and cut specific sequences, facilitating the precise manipulation of DNA.
DNA Ligase
An enzyme that joins DNA fragments through the formation of covalent bonds between them is DNA ligase. This enzyme is needed to seal the gaps that result from the insertion of a gene into vector DNA, to give a continuous DNA molecule.
Vectors
Vectors are DNA molecules responsible for transporting foreign genetic material into a host cell. Common vectors are plasmids, bacteriophages, cosmids, bacterial artificial chromosomes, and yeast artificial chromosomes. Every single one of the above-mentioned vectors finds application based on its unique features.
Host Cells
Host cells are the organisms into which recombinant DNA is introduced. Common hosts include bacteria, yeast, and mammalian cells, which offer a variety of advantages for the manipulation and expression of genes in these systems.
Steps in Recombinant DNA Technology
The steps for Recombinant DNA Technology are mentioned below:
Isolation of Genetic Material
Sources of DNA: It is possible to isolate genetic material from sources such as genomic DNA, cDNA, and synthetic DNA.
Methods of Extraction: The most common are cell lysis followed by membrane disruption and DNA purification, both by chemical reagents and kits specifically designed for a high yield and purity.
Diagram: DNA Extraction Process
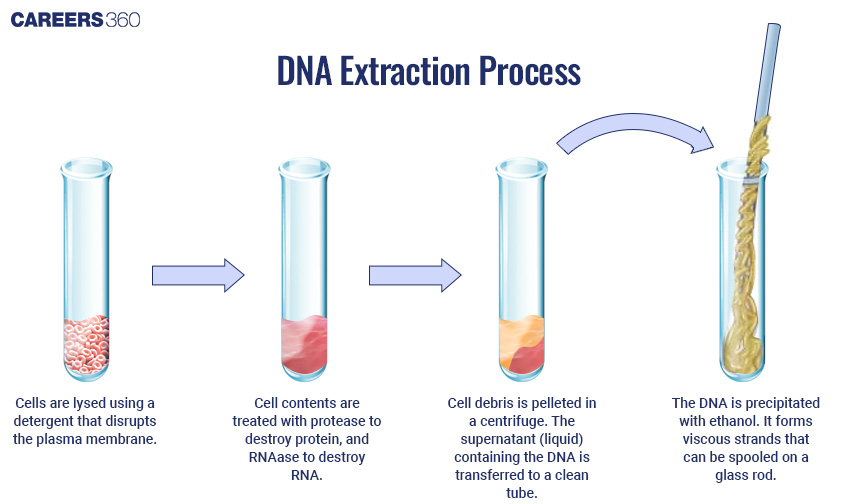
Cutting of DNA at Specific Locations
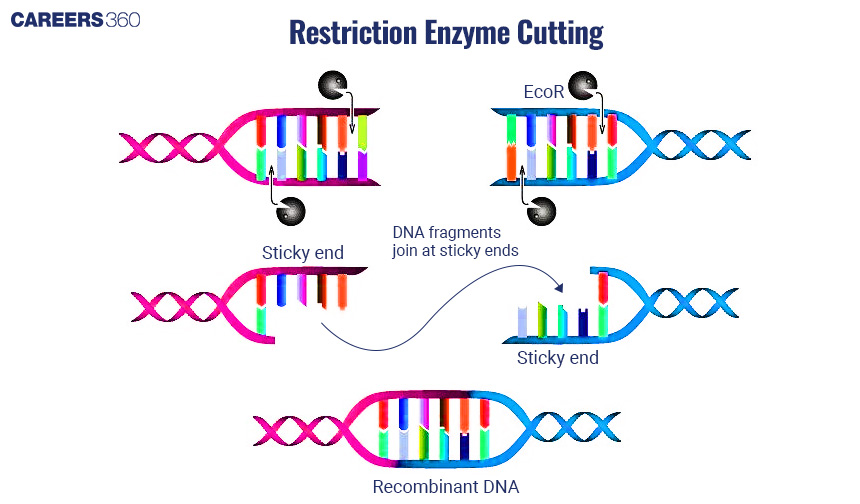
Use of Restriction Enzymes
These restriction enzymes cut the DNA at particular positions where the DNA sequences are recognized, thereby generating DNA fragments that could be manipulated for cloning.
Types of Restriction Enzymes
There are numerous such enzymes with diverse specificities towards DNA sequence recognition and cleavage. Among these are EcoRI and HindIII.
Diagram: Restriction Enzyme Cutting
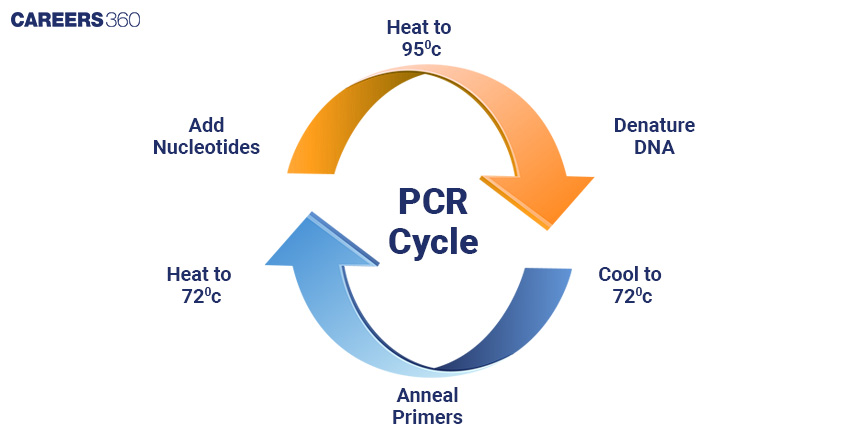
Amplification of Gene of Interest
Polymerase Chain Reaction (PCR)
PCR is a laboratory process that amplifies selected segments of DNA to make several million copies of a particular fragment of DNA.
Steps Involved in PCR
PCR is a process during which repeated cycles of denaturation, which is the separation of DNA strands, take place; annealing, the binding of primers to the DNA, and extension: synthesizing new DNA strands.
Diagram: PCR Cycle
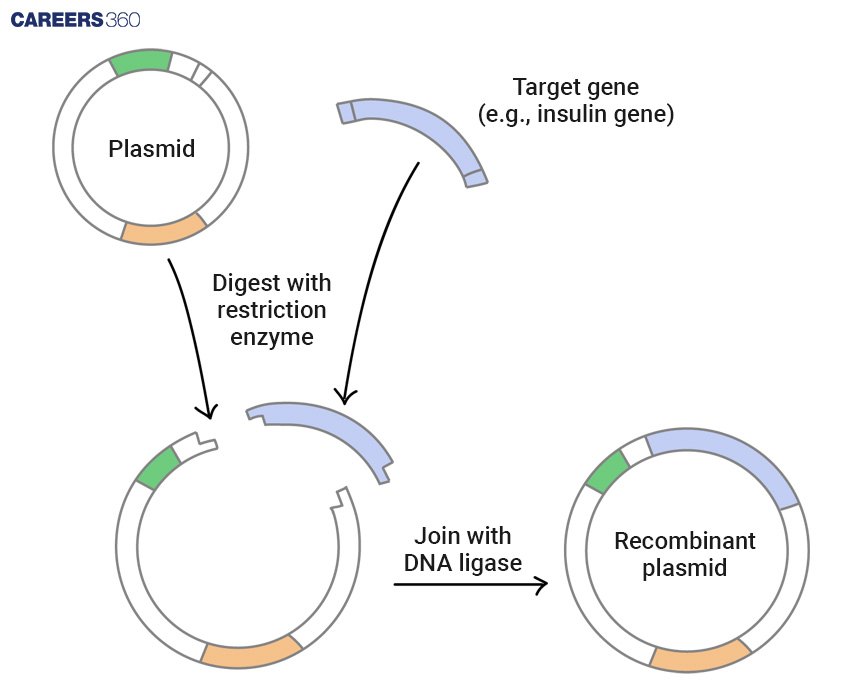
Inserting the Gene of Interest into a Vector
Types of Vectors
Examples of vectors that are included in recombinant DNA technology are plasmid, bacteriophage, cosmid, BAC, YAC, and the qualities and uses of each.
Ligation Process
A ligation is the process of attaching the amplified gene to a vector with the help of DNA ligase.
Diagram: Ligation of DNA into a Vector
Introduction of Recombinant DNA into Host Cells
Some methods of introduction are as follows:
Transformation Methods
Some methods that are used to introduce recombinant DNA into host cells include transformation; in bacteria, it is transfection; in eukaryotic cells and electroporation.
Selection and Screening of Transformed Cells
The next step is to examine transformed cells to ensure the host's uptake of recombinant DNA with the help of selectable markers and reporter genes.
Obtaining the Foreign Gene Product
Expression of the Recombinant Gene
The host cell expresses the recombinant gene resulting in the production of a protein or gene product of the desired type.
Methods of Protein Extraction and Purification
Extraction and purification of proteins are carried out with the help of various biochemical techniques in the host cells.
Applications of Recombinant DNA Technology
Medicine
Production of Insulin, Growth Hormones, and Vaccines
Human insulin, growth hormones, and vaccines have been created using recombinant DNA technology in medical science which has become a blessing for human beings suffering from diseases.
Agriculture
Development of Genetically Modified Crops
Genetically engineered crops that are modified for good traits in recombinant DNA technology include pest resistance, increased yield, enhanced nutrition, etc.
Industrial Biotechnology
Enzyme Production and Biofuel Development
Enzymes for industrial processes are produced using recombinant DNA technology and bio-fuels from renewable resources.
Environmental Biotechnology
Bioremediation and Waste Treatment
Bioremediation is also accomplished with the help of recombinant DNA technology by using genetically modified organisms to clean up environmental pollutants and waste.
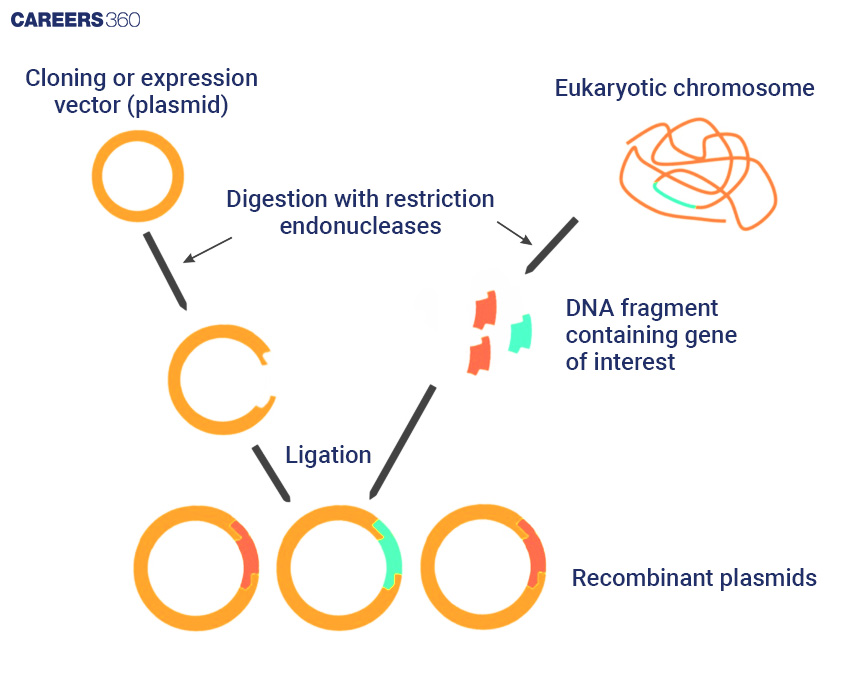
DNA Cloning
DNA cloning is the production of the identical copies of a DNA fragment. The process is composed of three steps: isolation of a fragment of DNA, insertion of the isolated DNA fragment into the vector, and introduction of the vector into the host cell.
Diagram: DNA Cloning Process
Vectors Used in DNA Cloning
Other vectors in common use are plasmids, bacteriophages, cosmids, bacterial artificial chromosomes (BACs), and yeast artificial chromosomes (YACs).
Applications of DNA Cloning
The DNA cloning technique is applied in the study of genes, used in medicine for the production of therapeutic proteins, and applied in agriculture for developing genetically modified crops.
Future Trends
Future trends are advances in personalized medicine, gene therapy, and the development of novel biotechnological applications.
Recombinant DNA technology has had an amazing impact on science and society. Its applications in medicine, agriculture, and industry have meant an upsurge in the relevance of these fields and promise further innovations. With technology growing rapidly, it is bound to bring discoveries and advancements that will shape the future of biotechnology.
Frequently Asked Questions (FAQs)
Recombinant DNA technology is the process of combining DNA from different sources to create a new genetic combination.
The steps are isolation of DNA, cutting of DNA using restriction enzymes, amplification with PCR, insertion into vectors, transformation into host cells, and obtaining the gene product.
Applications include insulin production, GM crop development, enzyme production, and bioremediation.
DNA cloning is the process of making numerous, identical copies of a DNA segment for various research, medical, and biotechnological uses.
Among the ethical considerations are impacts on health and the environment, genetic material patenting, and misuse of the technology.
Also Read
02 Jul'25 07:30 PM
02 Jul'25 05:51 PM
02 Jul'25 05:51 PM
02 Jul'25 05:50 PM
02 Jul'25 05:42 PM
02 Jul'25 05:42 PM
02 Jul'25 05:29 PM
02 Jul'25 05:29 PM